Given that the sharing of codon nucleotides dramatically reduces the size of protein sequences, viruses in nature often generate overlapping genes (OLG) in alternate reading frames of the same nucleotide sequence. The question of whether amino acid sequences are sufficiently degenerate with regard to protein folding to permit the extensive overlap of arbitrary pairs of functional proteins arises from their presence. Here, researchers use cutting-edge generative models to design synthetic OLGs to explore this subject. Researchers initially create overlapped sequences that target two distinct protein families to assess the strategy. After that, researchers encode unique, highly organized de novo protein structures and find that both in silico and experimental success rates are remarkably high. This shows that well-defined 3D folds in alternate reading frames can be accommodated simultaneously without being severely constrained by overlap limits imposed by the structure of the standard genetic code. According to our research, OLG sequences might be easily accessible in nature and could be used to compress and limit artificial genetic circuits.
Introduction
In 1977, Frederick Sanger solved a dilemma where the entire summed length of proteins produced by the bacteriophage ΦX174 was too large to fit into the calculated size of its DNA by sequencing its 5.4 kb DNA. Gene pairings are coded by the same section of DNA using distinct reading frames, yet previous observations were accurate. Because over half of all known viruses express at least one overlapping gene (OLG) in their genomes, OLGs have become a common occurrence, particularly in the field of virology. The number of potential OLGs across a broad range of cellular phylogenies has rapidly expanded due to advancements in mass spectrometry and ribosome profiling techniques. This has shown that many alternative open reading frames that are currently absent from reference gene annotation databases are expressed and functional.
Engineering OLGs with the CAMEOS Algorithm
The CAMEOS algorithm, which was created to integrate protein sequence direct-coupling analysis models, has demonstrated clear promise in the creation of synthetic OLGs for use in genetic biocontainment applications. This approach reduces the footprint of genes and payloads by rewriting genes to overlap with one another. The security of synthetic gene circuits is ensured by overlapping a gene of interest with an important selection gene, which also introduces containment measures and increases genetic stability. The active sites of Class I and II aminoacyl-tRNA synthetases were encoded using Rosetta in the first attempt at synthetic OLGs; Opuu et al. developed a more broadly applicable approach. Multiple synthetic OLGs with genetic biocontainment applications have been successfully constructed using the CAMEOS algorithm.
Key Aspects of the Research
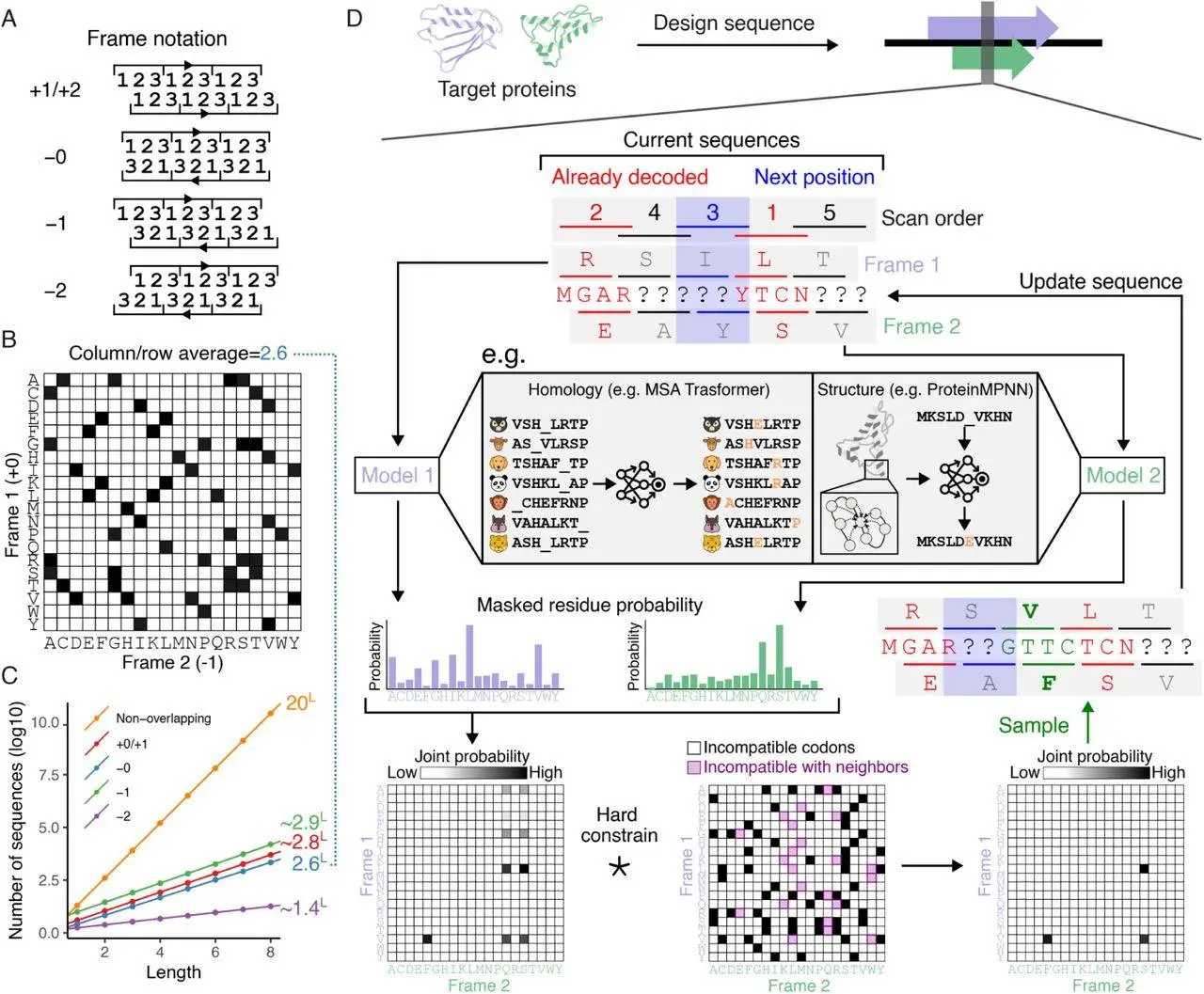
Researchers pondered whether using DL models could enhance the design of artificial OLGs. In this article, researchers present a computational technique that makes it possible to use the most advanced generative models of protein sequences available today for OLG creation. Researchers start by demonstrating a design scenario that focuses on two necessary and biosynthetic genes for entanglement conditioned on homologous protein families. Researchers demonstrate that although the resulting synthetic sequences differ greatly from natural sequences, they still perform comparably to them on various in silico benchmarks. Second, researchers create OLG sequences that, conditioned on the coordinates of their backbone atoms, encode pairs of highly ordered structures.
Experimental Outcomes of OLGs
It has become necessary to validate and evaluate the failure mechanisms of OLG designs due to the notable divergence in sequence composition. For recombinant expression and structural characterization, 192 overlapping sequences from an earlier design effort were chosen as a subset. Three pairs of proteins from various secondary structure groups were chosen for additional characterization out of the 207/384 (54%) individual proteins that the study found to have been successfully expressed. The success percentage varies depending on the secondary structure composition; proteins with only β sheets have a 30% success rate, whereas proteins with α helices have a 77% success rate. Some designs needed to be refolded from inclusion bodies, but the majority were purified from the cytoplasm. The total soluble yield was 8.5 mg/L of culture equivalent.
In OLG sequences, the success rate of overlapping proteins was found to be 31%, with 60/192 producing successful couples across all secondary structure classes. Success in a single frame has no systematic impact on the success rate because it is the same when looking at each protein separately. The solubility of recombinant proteins generated in E. coli was probably impacted by the OLG-specific compositional bias brought on by the frequent usage of high codon degeneracy amino acids, as seen by the substantial correlation between the success rate and the negative net charge of the proteins. OLG sequences encoding arbitrary pairings of well-defined 3D folds are feasible, despite the limited sequence space, as evidenced by the high experimental validation rates seen for non-overlapping sequence designs with ProteinMPNN.
Conclusion
Since their discovery in 1977, synthetic OLGs—which encode proteins in two distinct reading frames of the same nucleotide sequence—have posed a persistent biophysical and evolutionary conundrum. Under the restrictions of overlapping codons, a computational approach has been created to encode two target proteins into the same DNA sequence. A more thorough comprehension of protein sequences is made possible by this method, which builds on earlier work and integrates DL-based generative modeling of protein sequences. According to the study, stable protein backbones may be easily doubly encoded in different reading frames, which makes it very possible to construct synthetic OLGs using the methods currently used to generate protein sequences. However, there is a special difficulty in achieving effective opposite-strand OLG expression, which opens up a possibility for regulatory circuit design.
Article Source: Reference Paper | Code Availability: GitHub
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.