Scientists from the Earlham Institute and the University of Oxford have unexpectedly altered our understanding of the rules of genetics while testing a new single-cell sequencing method. A seemingly unique divergence in the genetic code signaling the end of a gene has been unveiled in the genome of a protist. This discovery suggests the imperative for additional research to understand this wide-ranging group of organisms better.
The genetic code, the language that dictates how DNA is translated into proteins, is one of the most conserved features across all life forms. However, nature always has a surprise in store. Among the myriad of species, a group called ciliates stands out for their intriguing genetic quirks. This article delves into the fascinating world of ciliates and explores how they have rewritten the genetic code, introducing variants rarely seen in other organisms.
The Universality of Genetic Code
The genetic code is like the universal grammar of life. It provides a blueprint for how the information stored in our DNA is read and translated into proteins. The code consists of three stop codons (UAA, UAG, and UGA) that signal the end of protein synthesis and 61 sense codons that correspond to specific amino acids. This canonical genetic code is employed by the majority of organisms on Earth.
Ciliates: Genetic Code Rebels
Ciliates are a unique group of single-celled eukaryotes. What sets them apart is their nuclear dimorphism, where each cell contains two distinct types of nuclei, the germline micronucleus (MIC) and the somatic macronucleus (MAC). The MIC serves as the genetic reserve, while the MAC takes the lead in other cellular functions.
Within ciliates, genetic code deviations are not unusual. These remarkable organisms have been found to redefine the meaning of certain codons, particularly the UAA and UAG stop codons. While for most species, these codons are synonymous, ciliates dare to be different.
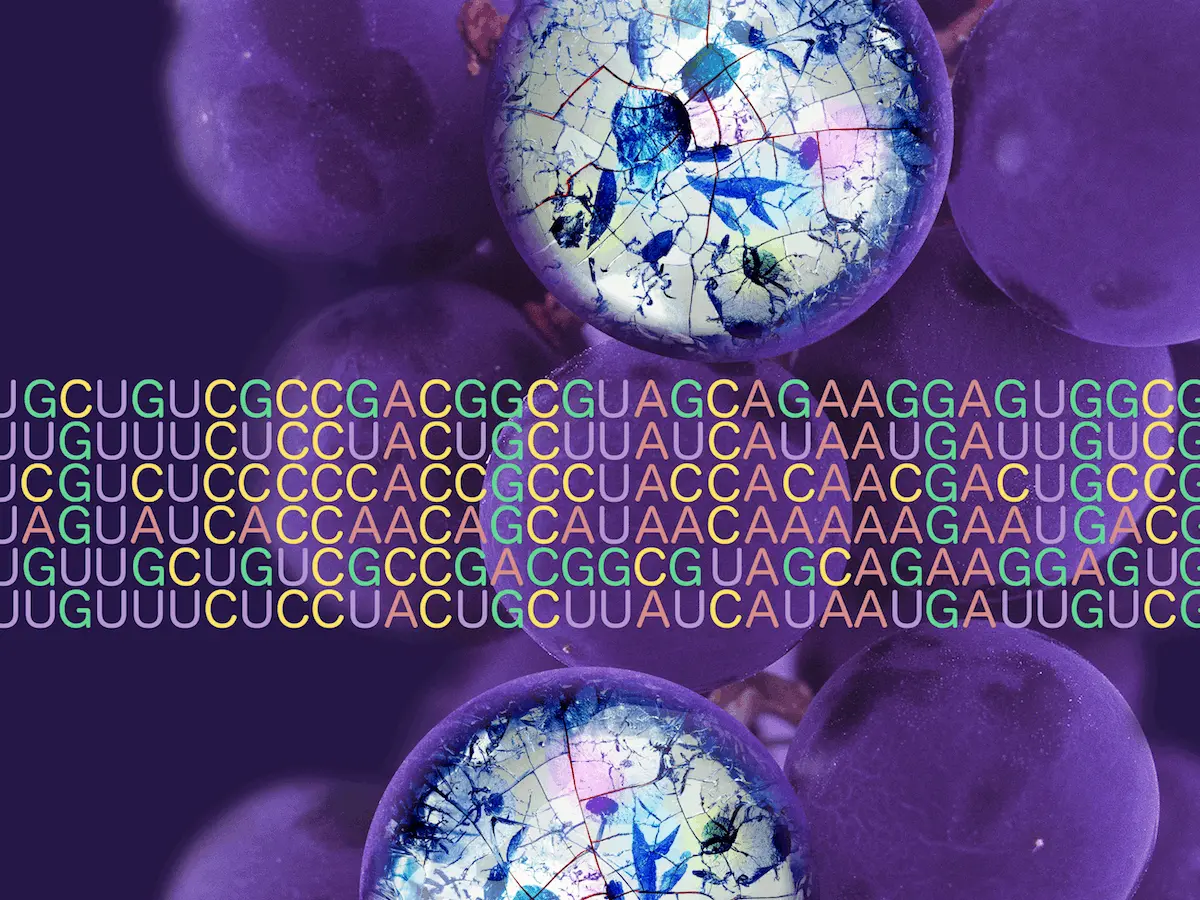
In a groundbreaking discovery, scientists have unveiled a novel uncultured ciliate belonging to the Oligohymenophorea class. The translation of UAA and UAG stop codons has been radically altered here. The UAA codon now encodes for lysine, while UAG has taken on the role of specifying glutamic acid. This revelation marks the first reported instance of UAA and UAG encoding different amino acids within a genetic code variant.
The Role of Suppressor tRNA Genes
Such a significant shift in genetic code functionality requires modifications to the translational machinery. In eukaryotes, the eukaryotic release factor 1 (eRF1) protein is responsible for recognizing standard stop codons. Mutations in the N-terminus of eRF1 can alter stop codon specificity, opening the door for genetic code reassignment.
In the event of translational readthrough, where the ribosome mistakenly continues protein synthesis past a stop codon, tandem stop codons come into play. These are additional stop codons strategically placed downstream of a gene. They act as a backup mechanism, minimizing erroneous protein elongation.
Several models have been proposed to explain genetic code variations. The “codon capture” model suggests that rare codons are gradually eliminated from the genome, only to reappear and be captured by a noncognate tRNA with a different amino acid. Alternatively, the “ambiguous intermediate” model envisions an intermediate stage where a codon is ambiguously translated by competing tRNAs or, in the case of stop codon reassignment, a suppressor tRNA competing with a release factor.
In the canonical genetic code, UAA and UAG typically have the same meaning, potentially due to wobble binding. This unique phenomenon allows a suppressor tRNA gene with a UUA anticodon to recognize both UAA and UAG codons. Experimental evidence in Tetrahymena thermophila supports this hypothesis.
In the case of Oligohymenophorea sp. PL0344, a ciliate species, the UAA and UAG codons have taken on distinct roles. While UAA now specifies lysine, UAG encodes for glutamic acid. This deviation from the norm showcases the adaptability and complexity of genetic codes in nature.
The genome of Oligohymenophorea sp. PL0344 boasts a rich collection of suppressor tRNA genes. Among them, tRNA-Sup(UUA) genes enable the recognition of both UAA and UAG codons. Similarly, tRNA-Sup(CUA) genes facilitate the translation of UAG into glutamic acid. These findings further validate the significant role of suppressor tRNA genes in genetic code reassignment.
The Oligohymenophorea sp. PL0344 genome assembly sheds light on other genomic features. With a genome size of approximately 100 megabases, it offers a comprehensive view of the genetic machinery that underpins this ciliate’s unique biology.
Implications for Evolutionary Biology
The discovery of this novel genetic code variant challenges established paradigms in evolutionary biology. It highlights the astounding adaptability of ciliates and raises intriguing questions about the co-evolution of stop codons and their corresponding release factors.
Conclusion
The genetic code is the cornerstone of life as we know it. Its universality binds all living organisms on Earth. Yet, within the enigmatic world of ciliates, we find a testament to nature’s penchant for diversity and innovation. The case of Oligohymenophorea sp. PL0344 opens a window into the intricate dance between genes, codons, and tRNAs, offering a glimpse into the ever-evolving narrative of life’s blueprint.
As we continue to explore the genetic codes of diverse organisms, we may uncover further surprises, each one adding a new layer of complexity to the story of life on our planet. The study of genetic code variants in ciliates reminds us that there is still much to learn and that the microscopic world holds untold secrets waiting to be revealed.
Story source: Reference Paper
Learn More:
Dr. Tamanna Anwar is a Scientist and Co-founder of the Centre of Bioinformatics Research and Technology (CBIRT). She is a passionate bioinformatics scientist and a visionary entrepreneur. Dr. Tamanna has worked as a Young Scientist at Jawaharlal Nehru University, New Delhi. She has also worked as a Postdoctoral Fellow at the University of Saskatchewan, Canada. She has several scientific research publications in high-impact research journals. Her latest endeavor is the development of a platform that acts as a one-stop solution for all bioinformatics related information as well as developing a bioinformatics news portal to report cutting-edge bioinformatics breakthroughs.