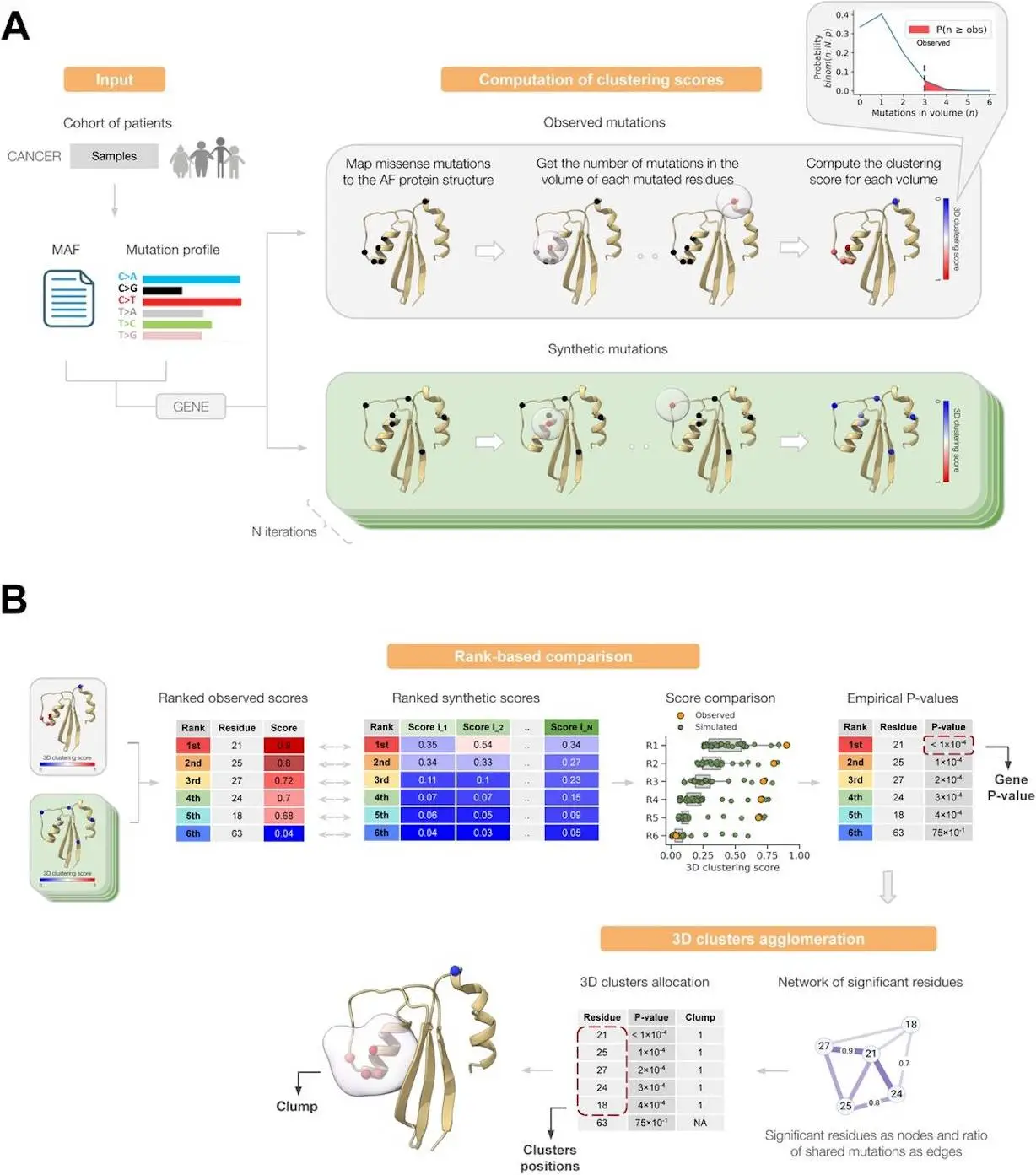
Finding the genes that can cause tumors in various tissues is one of the main objectives of cancer genomics. Computational techniques that use various positive selection signals in the pattern of gene mutations seen in tumors have been created for this goal. One indication of positive selection is the accumulation of mutations in regions of the protein’s three-dimensional (3D) structure that are higher than predicted in a neutral state. Proteins with experimentally determined 3D structures that cover their whole sequence are scarce, which has made it difficult to develop methods that take advantage of this signal. Here, researchers from IRB Barcelona introduce Oncodrive3D, a computational technique that uses AlphaFold 2 structural models to identify proteins with significant mutational 3D clusters across the entire human proteome. Oncodrive3D outperforms state-of-the-art techniques for identifying cancer driver genes that use mutational clustering in terms of computational efficiency and exhibit sensitivity and specificity comparable to those techniques. Researchers provide multiple examples illustrating how the significant mutational 3D clusters discovered by Oncodrive3D in various known or putative cancer driver genes can provide information about the tumorigenesis mechanism in various cancer types and clonal hematopoiesis.
Introduction
Cancer genomics is concerned with identifying mutational cancer driver genes that can cause tumorigenesis in response to point mutations and indels. Because somatic point mutations are available across tumor cohorts, computational methods have been developed to compare mutation features with neutral mutagenesis, such as the distribution of mutations along the sequence or protein structure, the average functional impact, and the recurrence of mutations, significant differences between the observation and expectation under neutrality are referred to as signals of positive selection. The values of these features in various cancer genes reveal information about the underlying mechanisms of tumorigenesis, such as gain-of-function genes accumulating missense mutations at specific positions and loss-of-function genes accumulating more truncating mutations along their sequence than expected.
Understanding Oncodrive3D
Oncodrive3D is a quick and precise new 3D clustering approach for identifying cancer driver genes. Oncodrive3D uses predicted aligned error (PAE) and AlphaFold 2 (AlphaFold for short) structure predictions to create contact probability maps and find mutational clusters in the three-dimensional protein structure across all altered genes in a tumor cohort. To achieve this, it uses rank-based statistics to calculate empirical p-values to identify residues displaying a mutation accumulation in their 3D structural vicinity that is noticeably greater than that predicted under neutrality, as well as the trinucleotide mutational profiles are seen across tumors to simulate neutral mutagenesis. Next, Oncodrive3D deduces clusters of residues in the protein’s three-dimensional structure with a notable accumulation of mutations.
In an investigation including 28,067 tumors representing 83 different types of cancer, Oncodrive3D outperforms a cutting-edge 3D clustering-based driver discovery technique, expanding the discovery to the complete human proteome while consuming 33 times fewer CPU days. To help the methodical effort to finish compiling the list of mutational cancer driver genes, it works in conjunction with other driver discovery techniques based on various feedback loops. Oncodrive3D additionally offers comprehensive annotations of mutational clusters that arise during positive selection in the three-dimensional structure of cancer proteins.
Applications and Performance
- Oncodrive3D detects notable differences between the clustering of mutations in a protein’s 3D structure seen in different tumors and what would be predicted in the same group of tumors if the protein were neutral. This is accomplished by scoring and comparing the local accumulation of observed mutations in three-dimensional (3D) space (3D clustering score) at each mutant amino acid residue in three-dimensional space to that calculated for synthetic mutations produced after the neutral mutagenesis process.
- Oncodrive3D shows comparable performance to another state-of-the-art mutational 3D clustering method and outperforms a linear mutational clustering method. The reliance of Oncodrive3D on AlphaFold models guarantees a maximum coverage of all potential cancer driver genes with significant mutational 3D clusters.
- Oncodrive3D has a higher computational efficiency than mutational clustering-based driver discovery techniques. It shows comparable performance to techniques that do not rely on working with 3D protein structures and instead take advantage of signals of positive selection.
- A comparison of mutational 3D clusters in DNMT3A in CH and those in AML cases showed that significant residues within the methylase domain are more recurrent than those in other areas of the protein. Oncodrive3D has been used to identify genes with significant mutational 3D clusters in clonal hematopoiesis (CH), the normal expansion of clones in the blood. Oncodrive3D identified several well-established CH driver genes by analyzing somatic mutations in blood from 36,461 donors throughout three cohorts.
Conclusion
Oncodrive3D is a computational technique that finds genes that can drive carcinogenesis by identifying genes with notable mutational 3D clusters across tumors. Contributing to identifying driver mutations across cancer genes, this approach offers insights into their carcinogenesis mechanisms in various tissues. Oncodrive3D performs better in terms of computational resource economy than other clustering-based driver-finding techniques. It complements other positive selection signals and exhibits sensitivity and specificity comparable to other techniques. The technique is intended to be incorporated into the intOGen pipeline, which runs on cohorts of tumors gathered from the public domain and aggregates the results of seven driver-finding techniques. It may be possible to expand Oncodrive3D to detect mutational 3D clusters at the interface of interacting proteins.
Article Source: Reference Paper | Oncodrive3D is available on GitHub.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.