Cancer, a complex disease arising from genetic alterations, has been a major global health challenge. Researchers have been able to make considerable progress in their understanding; however, the poor availability of real-world data on the cancer genome has limited the progress. To overcome this loss, a group of researchers at the Ontario Institute of Cancer Research and the University of Toronto led by Ander Díaz-Navarro and Lincoln Stein devised a new methodology to create synthetic cancer genomes by means of generative AI.
This method, OncoGAN, extends chances to cancer researchers by offering ample and various types of realistic synthetic cancer genomes. Because of the unencumbered data privacy restrictions, OncoGAN provides opportunities for researchers to create and improve methods that will result in better cancer diagnostics, prognostics, and treatments.
The Role of Synthetic Data
Synthetic data is created by algorithms and has the potential to solve the problem. By mimicking real-world data while preserving privacy, it creates possibilities for scientists to analyze practically big and diverse databases. In relation to cancer studies, synthetic genomes of cancer cell lines could facilitate breakthroughs in the development of precision oncology concepts.
Introducing OncoGAN
OncoGAN is a cutting-edge generative AI tool that will produce highly realistic-looking synthetic cancer genomes. Such a tool is solving the problem of real-world data scarcity by providing large-scale privacy-compliant and diverse datasets at the click of a button.
How OncoGAN Works
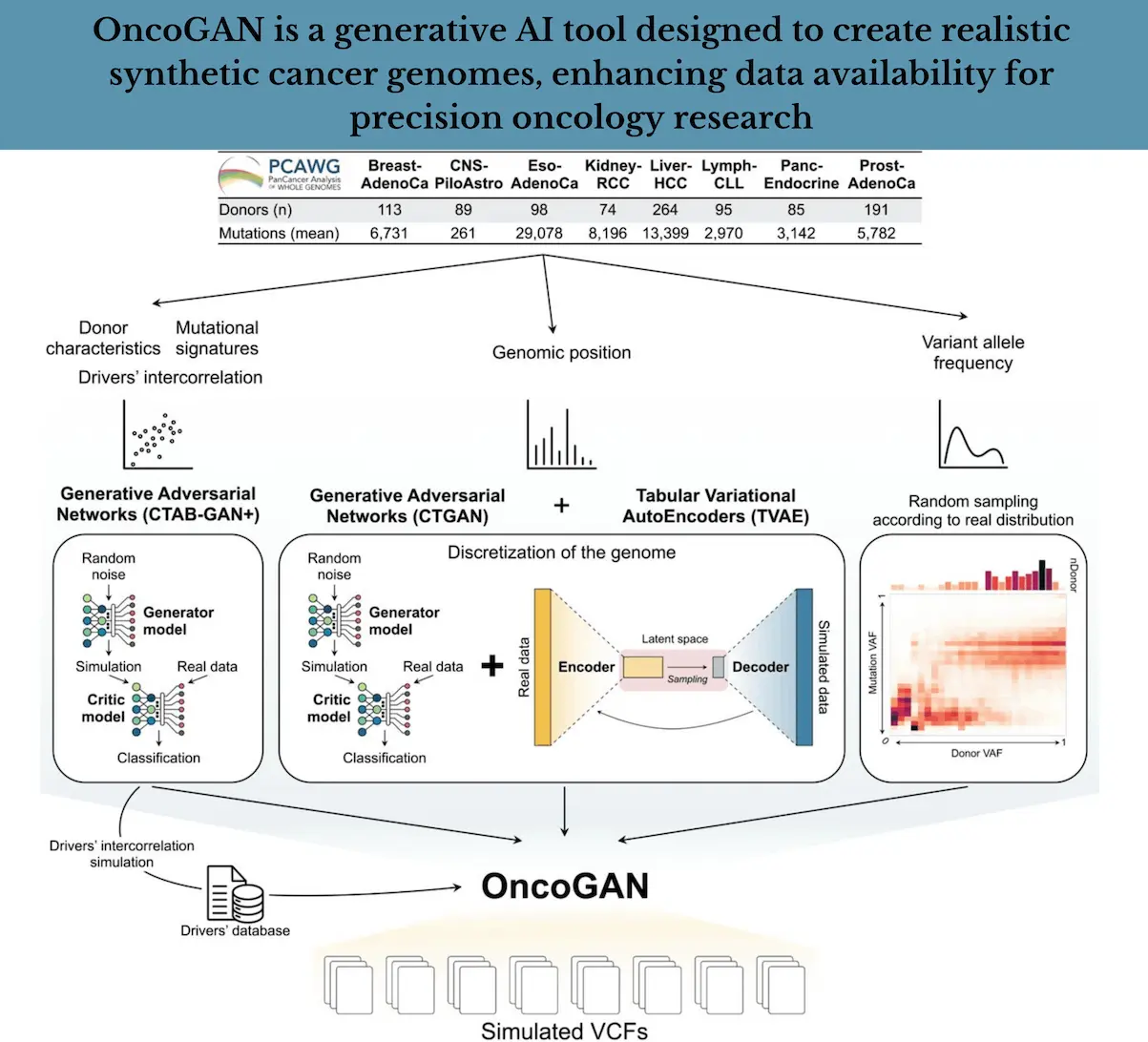
The ingenious artificial intelligence technique OncoGAN employs several strategies to achieve the goal of producing synthetically made cancer genomes and does so beautifully.
Let us elaborate on the key features of OncoGAN:
Data Preprocessing:
- Real-world data: The foundation of OncoGAN lies in real-world cancer genome data, sourced from large-scale genomic projects like the International Cancer Genome Consortium (ICGC).
- Feature extraction: This researcher focused on the processing and extraction of significant features—types of mutations, their position on chromosomes, and frequencies of variation.
- Data cleaning and normalization: The data thus obtained also undergoes some cleaning and purification to maintain consistency and is also normalized to match the requirements of the training of the model.
Model Training:
- Generative Adversarial Networks (GANs): The use of GANs involves generating simulated data that closely resembles real data. For OncoGAN, researchers have trained GANs on the cancer genome data that has been preprocessed to learn the patterns and produce realistic made-up genomes.
- Variational Autoencoders (VAEs): Other powerful generative deep learning techniques meant to learn a latent representation of the data are VAEs. However, in the case of OncoGAN, VAEs capture the complex relationships between different genomic features and generate diverse synthetic genomes.
Synthetic Genome Generation:
- Mutation Simulation: OncoGAN is also capable of generating a wide variety of mutations, including SNPs, insertions, deletions, and structural variations. Such mutations are derived from the patterns learned from real data.
- Mutational Signature Simulation: This tool is able to reproduce the characteristic mutational signatures associated with different types of cancer, as well as the biological processes behind them.
- Driver Mutation Simulation: Driver mutations, which directly contribute to cancer development, are incorporated into the synthetic genomes. The model’s possible mutations are made to take into account the frequency of such mutations and the patterns of their co-occurrence.
- Variant Allele Frequency Simulation: Variant Allele Frequency simulation features the accurate simulation of allele variant frequencies in a sample concerning real-world data variability.
- Genomic Position Simulation: The location of mutations within the genome is carefully simulated to reflect the observed patterns in real cancer genomes.
Evaluation and refinement
- Quality Control: The generated synthetic genomes are rigorously evaluated against real-world data using various metrics, including:
- Mutational signature profiles
- Driver mutation patterns
- Variant allele frequencies
- Genomic location of mutations
- Model Refinement: The model continuously undergoes various refinements based on the evaluation to enhance the accuracy and realism of the synthetic genomes.
Combining these techniques, OncoGAN produces synthetic cancer genomes that closely mimic the complexity and diversity of real-world cancer data. This amazing tool can change the face of cancer research as it provides researchers with a necessary tool in the quest for new therapeutics as well as a more detailed insight into the disease.
Conclusion
OncoGAN is a development that should be celebrated in the field of cancer research. In particular, this tool addresses the challenges brought about by the availability of real-world data and provides a valid artifact that can aid researchers in progressing research in a fraction of the time taken previously. As AI continues to advance, we can expect even more sophisticated tools to emerge, further revolutionizing our understanding of cancer and our ability to combat this disease.
There is a great deal of optimism regarding the future of cancer research, and undoubtedly synthetic data, as shown through OncoGAN, will aid this direction. Using AI, new avenues will be found, newer and better treatments will be conceived, and ultimately improve the lives of cancer patients.
Article Source: Reference Paper | The OncoGAN software is publicly available on GitHub and DockerHub to enhance reproducibility.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.