Combination therapies have emerged as a new strategy in the battle against cancer and are viewed as superior to previously existing treatment options. There is a possibility that multiple pathways involved in cancer progression may be targeted that may foster overcoming the usual resistance that comes with single-drug therapies. However, the scope of drug combinations tends to make it prohibitively difficult to test all of the many possible combinations. The number of potential pairings rises to several tens of millions with only 206 pairs of distinct drugs and a single approach, highlighting the impractical nature of traditional screening methods.
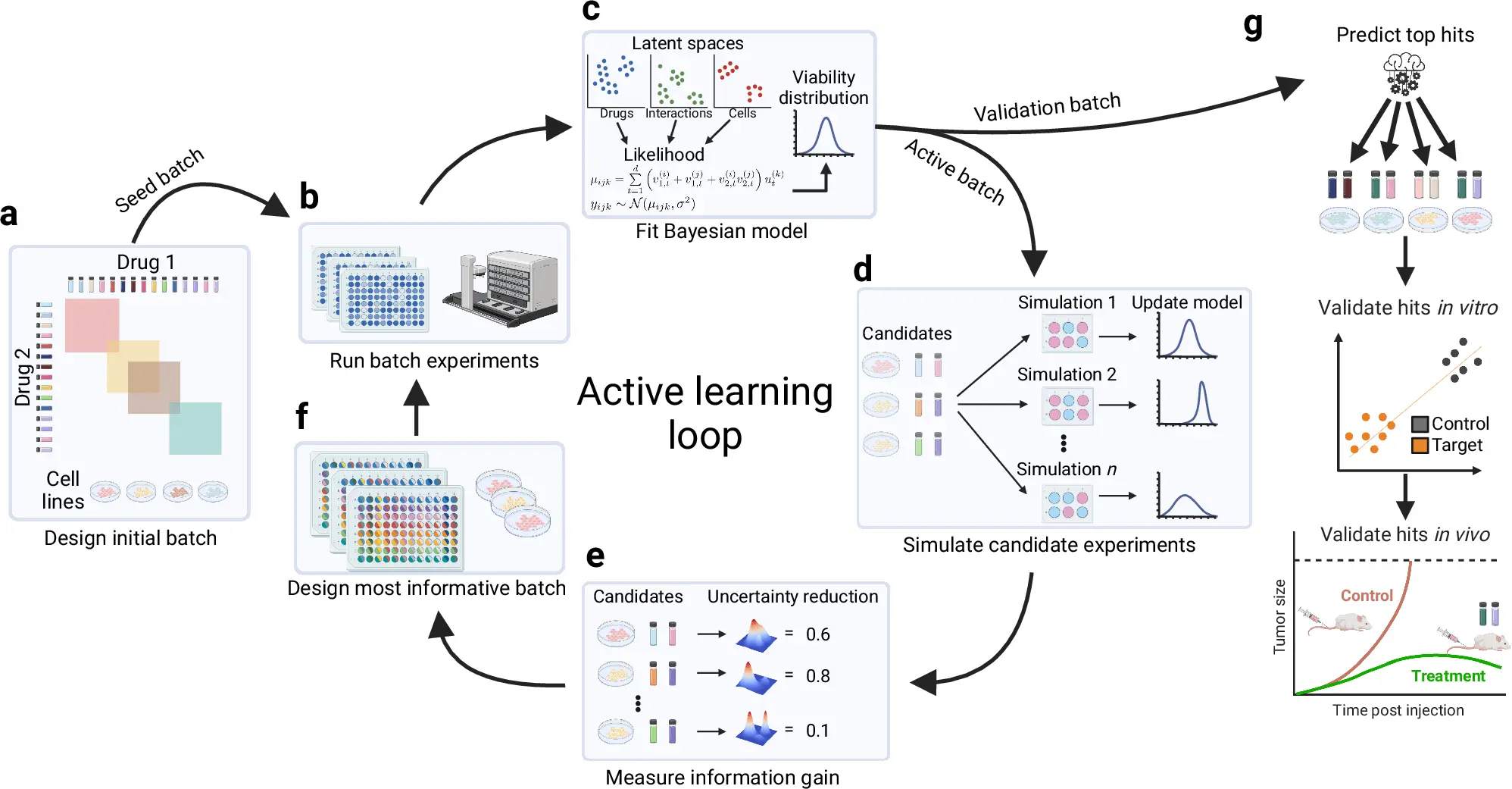
This is where the revolutionary approach of BATCHIE comes in, offering a dynamic and efficient method for large-scale combination drug screening. Developed by researchers at Memorial Sloan Kettering Cancer Center (MSKCC), BATCHIE represents a paradigm shift in how we approach drug combination experiments, and its potential is transforming the landscape of cancer research.
The Problem with Traditional Drug Screening Methods
In traditional combination drug screening, researchers measure the potency of a combination of predefined pairs in potentially blocking the growth of cancer cells in cell lines. Given the huge number of possible combinations, it is impractical to screen all possible drug pairs in detail, especially for large compound libraries. Similar to existing methods. However, these attempts also suffer from a limitation that is quite straightforward: They are bound to rigid experimental setups and, therefore, could not find important interactions or synergies between drug pair combinations.
Introducing BATCHIE: A Dynamic and Efficient Approach
BATCHIE is an acronym for BATCH Integrated Experimentation. It approaches Batch Experimentation with a dynamic angle by performing experiments in batches. Rather than trying out every possible combination of the drugs beforehand, BATCHIE leverages an active learning algorithm that chooses only the most useful experiments based on the previous ones conducted.
This design is bipartite and probabilistic in its orientation and reminiscence of information theory. In all given cases, the interface observes the previous tests performed and selects and recommends several combinations that are most likely to yield useful results. This approach ensures that the significant effort invested in the experiment has not been wasted – any tests that promote the identification of suitable drug pairs are encouraged.
The Results: More Synergies with Fewer Experiments
To validate the power of BATCHIE, the researchers applied it to retrospective experiments from previous large-scale combination drug screens. The findings were shocking since BATCHIE was capable of discovering potent and synergistic drug combinations almost instantaneously by identifying the most suitable combinations to test next. This demonstrates how adaptive experimentation assisted by probabilistic models can be beneficial in the drug screening process.
BATCHIE was put to challenge while in action as part of the true integration combination experiment, in which it could predict the effectiveness of a combination of an unprecedented 1.4 million combinations of a library composed of 206 drugs with a plethora of pediatric cell lines. Surprisingly, even with such a vast untreated combination, the BATCHIE model learned quickly, enabling it to predict new combinations that had not yet been designed. Moreover, one of the combinations that BATCHIE was able to identify was that of PARP inhibition plus topoisomerase I inhibition for the treatment of Ewing sarcomas. This combination was rational and later substantiated through follow-up validation experiments.
A Game-Changer for Ewing Sarcoma Research
There is great potential in BATCHIE’s capacity for selecting a drug combination panel for Ewing sarcoma – an aggressive form of cancer that occurs in children and young adults. Drug combination experiments were able to provide some candidate drug combinations for further development. All of these predicted drug combination efficacies were validated during experiments, thereby confirming BATCHIE’s usefulness in developing actionable therapeutic strategies.
As emaciated, hard-to-reach cancer targets become characterized, some of the most suitable pairing for Ewing sarcoma emerged as a combination of PARP inhibition plus topoisomerase I inhibition, indicating that BATCHIE can locate new synergies as well as rank combinations with clinical benefit.
The Advantages of BATCHIE
The most important benefit of BATCHIE is the reduction of combinatorial experiments, which allows carrying out almost unlimited drug rescans without primary screening. By concentrating only on the most informative augmentations, BATCHIE minimizes the amount of screening required, which translates into considerable savings in time, resources, and workforce. In actuality, BATCHIE was able to pinpoint excellent drug combinations after conducting tests on very few of the possible pairings, just 4 percent of the 1.4 million possible drug combinations in the job of screening for pediatric cancer drugs.
Additionally, BATCHIE, being an unbiased platform, is not restricted by any assumptions made a priori or exponents constraints. The adjustable applicability of BATCHIE guarantees that every single research study aids in reconstructing the knowledge behind drug interaction and its effect on the drug synergy continuum. This provides a scope for further research as BATCHIE does not limit itself to the already existing interactions but encourages the formation of new ones, which could potentially be entirely novel.
Conclusion: A New Era in Cancer Research
BATCHIE combines three key aspects of today’s research – theory of information, modeling of probabilistic outcomes, and dynamic designing of the experiments. The ability to predict synergistic drug combinations with fewer experiments not only leads to faster drug discovery but also shatters the limitation of the traditional one-size-fits-all approach to drug development, resulting in more effective treatment in the case of cancer.
With its open-source availability and proven success in identifying new therapeutic strategies for cancers like Ewing sarcoma, BATCHIE is poised to make a lasting impact on the field of cancer research. By reducing the cost and complexity of combination drug screening, BATCHIE offers a faster, more efficient route to discovering life-saving cancer therapies. The future of cancer treatment may very well be shaped by adaptive, information-driven approaches like BATCHIE.
Article Source: Reference Paper | BATCHIE is open source and available publicly on GitHub.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Follow Us!
Learn More:
Anchal is a consulting scientific writing intern at CBIRT with a passion for bioinformatics and its miracles. She is pursuing an MTech in Bioinformatics from Delhi Technological University, Delhi. Through engaging prose, she invites readers to explore the captivating world of bioinformatics, showcasing its groundbreaking contributions to understanding the mysteries of life. Besides science, she enjoys reading and painting.