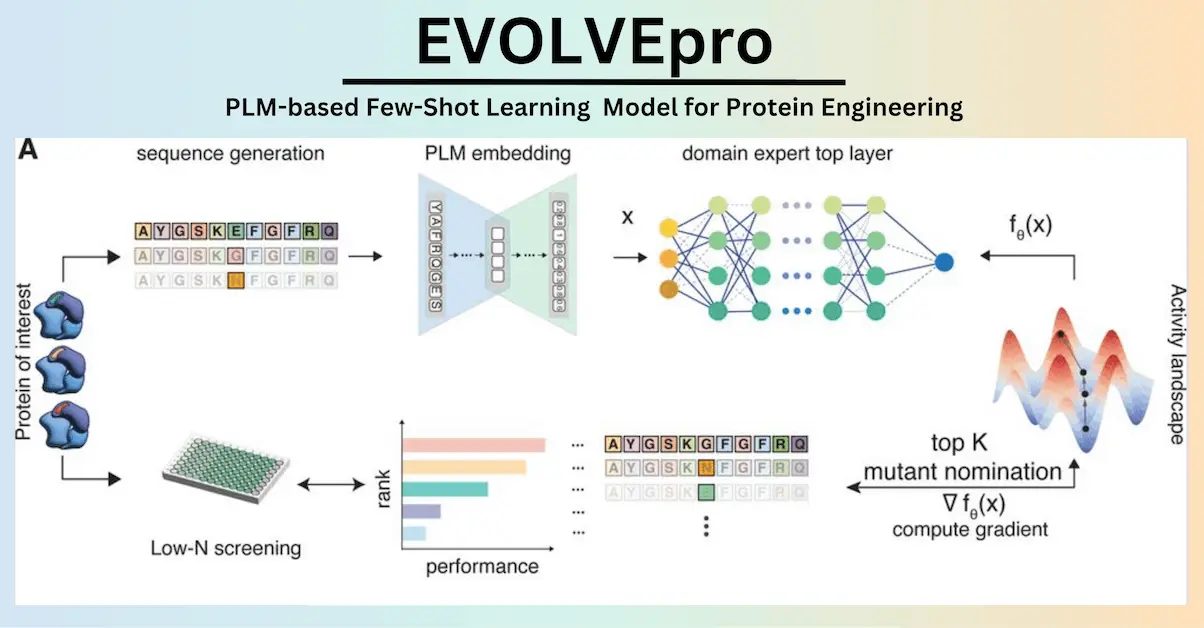
The use of directed evolution techniques is essential for sustainability, medicines, and protein research. These techniques, however, struggle to optimize several attributes and are labor-intensive. Though present approaches do not generalize across varied protein families, silicon-directed evolution methods using protein language models have the potential to expedite innovation. Researchers present EVOLVEpro (EVOlution Via Language model-guided Variance Exploration for proteins), a few-shot active learning system that combines PLMs and protein activity predictors to rapidly enhance protein activity, delivering improved activity in as few as four rounds of evolution. EVOLVEpro significantly improves in silico protein evolution, enhancing desired properties by up to 100-fold. It is used in three applications: T7 RNA polymerase for RNA production, a miniature CRISPR nuclease, a prime editor, an integratase for genome editing, and a monoclonal antibody for epitope binding. These results demonstrate the advantages of few-shot active learning over zero-shot predictions, paving the way for broader AI-guided protein engineering in biology and medicine.
Introduction
Protein diversity is an essential component of biological evolution, and deep learning has the capacity to transform it completely. Protein language models (PLMs) can be trained to complete masked amino acids across extensive protein sequence databases, therefore acquiring the grammar of protein variety. Although there are evolutionary restrictions and a limited amount of training data, these models can be utilized to identify protein variations with increased activity; nonetheless, their generalization to novel situations is limited. Active learning techniques have improved a variety of proteins but at the expense of thorough experimental examination. One such technique is machine learning-directed protein evolution (MLDE). Active learning and PLMs together might make the evolution process easier and get around this restriction, but attempts to do so have not progressed past proof-of-concept experiments such as fluorescent protein engineering.
Understanding EVOLVEpro
Researchers in this study present a novel protein evolution model, EVOLVEpro. The learning framework EVOLVEpro evolves high-activity mutants by utilizing protein sequences. The method employs a modular design, merging a top-layer regression model and an evolutionary-scale PLM to determine the activity landscape of a protein. As a result, protein sequences with increased activity can now be generalized. Because of its modular design, the framework simply needs protein sequences; structural knowledge or previous data are not required. This allows it to grow with larger parameter PLMs. Abundant opportunities in biology and medicine arise from EVOLVEpro’s capacity to evolve numerous protein functions simultaneously.
Methodology
The state-of-the-art performance of the protein language model EVOLVEpro has been demonstrated by benchmarking across 12 protein datasets. Three applications of the model were used: mRNA production, genome editing, and antibody medicines. In addition to demonstrating in vivo liver editing using an EVOLVEpro-engineered tiny nuclease and better in vivo mRNA performance from an EVOLVEpro evolved T7 polymerase, EVOLVEpro produced mutants with improvements ranging from 2 to 515 times over the original proteins. Additionally, the model investigated several residues in unexpected protein positions, exposing a unique activity landscape that frequently had a negative correlation with the fitness landscape deduced by the underlying protein language model. Because of EVOLVEpro’s ability to generate several mutant protein sequences from a large sequence space, final mutants with significantly higher activity than naturally occurring proteins are made possible. This demonstration demonstrates the capabilities of few-shot active learning with protein language models for optimizing proteins for diverse activities.
Application
Researchers have evaluated five therapeutically important proteins using EVOLVEpro, a unique AI protein engineering model. A number of attributes may need to be optimized simultaneously for these proteins, as evidenced by the poor connection seen between their activity and PLM-estimated fitness. With more than 16,000 potential sequences and more than 780 billion sequences to choose from, EVOLVEpro can identify highly active single mutants and multi-mutants. In order to determine SOTA performance, the models have been verified for tasks including genome editing, binding, and RNA synthesis. The enhancement of activity through various methods is revealed by a structural study of top mutations, indicating potential avenues for enzyme development in the future. EVOLVEpro is highly capable in protein design, as it has high success rates, requires no special knowledge about the protein, can be used for multi-objective function optimization, and is highly modular. The model is expected to improve with new foundation models and enhanced search strategies, making it broadly useful for protein engineering.
EVOLVEpro: Advantages, Setbacks, and Future Opportunities
The intrinsic drawbacks of PLMs have been effectively addressed by the generative PLM EVOLVEpro, which is trained to perform sequence reconstruction tasks throughout evolutionary variation. Nevertheless, the learnt fitness landscape of the PLM might not coincide with the activity landscape of a protein, and natural sequences do not always choose the best protein activity. PLMs can scale with increasing parameters, but enzyme optimization is more complex, and new research has revealed saturation scaling effects on bigger models. Although functional de novo proteins like CRISPR nucleases and GFP have been produced by generative PLMs, the functional success rate of these proteins is modest, and their variants do not exhibit enhanced activity compared to wild-type proteins. Future generative PLMs with improved architectures and training data may be suitable for combining with EVOLVEpro to create a design framework for rapidly optimizing de novo generated sequences for activity.
Conclusion
The ensemble model EVOLVEpro evolves protein activity through few-shot active learning. With just a few cycles of evolution, it creates highly active mutants by learning broad laws governing protein activity using evolutionary scale PLMs and a regression layer. Because of its extensive latent space and robust feature selection, EVOLVEpro is a low-N learning method that necessitates little wet lab testing. EVOLVEpro outperforms conventional encoding techniques like one-hot encoding and integer encoding, as demonstrated by benchmarking it against 12 distinct DMS datasets that span eight protein classes. Because of its modular nature, EVOLVEpro is excellent in low-N evolution scenarios since it can incorporate future advancements into autoregressive PLMs. Future advancements in autoregressive PLMs can be integrated because of the modular design.
Article Source: Reference Paper | Models and codes are available in the following GitHub repositories: Full data repository and Deployable model.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.