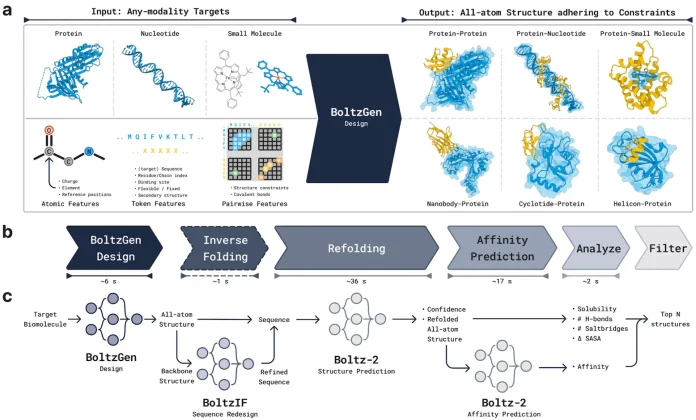
Researchers at MIT introduce an all-atom generative model, BoltzGen. The model incorporates a design specification language that allows researchers to control constraints like covalent bonds, binding sites, and structural motifs for designing proteins and peptides that can bind to diverse biomolecular targets. With strong structural reasoning and state-of-the-art folding performance, BoltzGen is experimentally validated across multiple wet-lab campaigns, advancing protein design and therapeutic discovery.
Bridging Binder Design and Structure Prediction
Traditional drug discovery is a slow and expensive process, often taking 10-15 years and billions of dollars, with a high likelihood of failure. Generative models can rapidly propose candidate binders like proteins or peptides, cutting down the number of wet-lab experiments needed. Though earlier approaches utilized models that are specialized for one type of biomolecule (e.g., nanobodies only, or peptides only). Also, other models typically separate the tasks of binder designing and structure prediction (like Alphafold). This limits their usefulness when we want a general tool that can design across modalities.
BoltzGen is positioned as a solution to such limitations as it works across nanobodies, peptides, proteins, and even small molecules. It also combined the task of designing binders and predicting their fold structures simultaneously.
Understanding How BoltzGen Treats Binder Design and Structure Prediction as One Problem
Core Idea: A Single All-Atom Diffusion Model
Diffusion model is a type of generative AI that starts from random noise and gradually refines it into a structured output (here, a protein or peptide structure). Unlike models that only represent backbone atoms, BotzGen explicitly models every atom in the molecule. This allows it to capture fine-grained interactions like hydrogen bonds, salt bridges, and side chain packing.
What sets BoltzGen apart is that if we give it only the sequence of a target protein, it will fold the target and design a candidate binder at the same time, producing a bound complex. This unified approach reduces errors and ensures consistency between design and folding.
Geometric Encoding of Residues
Instead of labeling residues with discrete categories, Boltzen encodes them geometrically. Each design residue is represented by a fixed set of atoms. The models learn to place these atoms in specific positions relative to the backbone. The arrangement of these atoms signals the identity of the residue. This keeps everything in a continuous geometry, which is ideal for diffusion models. It also helps to avoid mixing discrete labels with continuous coordinates, making the training scalable and efficient, similar to AlphaFold3.
Wet Lab and Computational Validation Results
In wet lab experiments, BoltzGen’s designs were tested across eight major experimental campaigns covering nanobodies, full proteins, linear peptides, cyclic disulfide-bonded peptides, anti-microbial peptides, and even small molecules.
Most notably, the team targeted nine completely novel proteins with no close bound structures in PDB and achieved nanomolar binders for about two-thirds of them, using both nanobody and protein formats.
Only a small number of designs were experimentally tested, yet meaningful binders were repeatedly discovered, showcasing strong generalization.
In computational validation results, its folding accuracy is on par with Boltz-2, which is a state-of-the-art structure prediction model. The test was carefully curated by clustering sequences at a 40% similarity threshold (to avoid redundancy). About 187 complexes were excluded because they didn’t fit into GPU memory, but otherwise, BoltzGen matched Boltz 2’s folding results.
Limitations of BoltzGen
High-affinity binders are necessary but not sufficient for drug development. A therapeutic binder must also have selectivity, developability, and target-specific considerations. So BoltzGen’s success in generating string binders is only the first step toward a viable drug.
Authors specifically highlight the memorization issue, that BolzGen sometimes memorizes ubiquitin when designing binders of length 73 to 76. Hence, ubiquitin is overrepresented in the PDB (>900 entries). This causes diversity to collapse, and the model repeatedly samples ubiquitin-like binders.
Conclusions and Final Takeaways
BoltzGen is positioned as the first truly general-purpose, open-source generative model for binder design, validated across diverse modalities and targets, and released as a complete pipeline for the scientific community. It’s not just a model but an end-to-end pipeline that could accelerate therapeutic discovery, biosensor development, and synthetic biology by providing a universal design engine.
Article Source: Reference Paper | Reference Article | Code availability: GitHub
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Saniya is a graduating Chemistry student at Amity University Mumbai with a strong interest in computational chemistry, cheminformatics, and AI/ML applications in healthcare. She aspires to pursue a career as a researcher, computational chemist, or AI/ML engineer. Through her writing, she aims to make complex scientific concepts accessible to a broad audience and support informed decision-making in healthcare.















