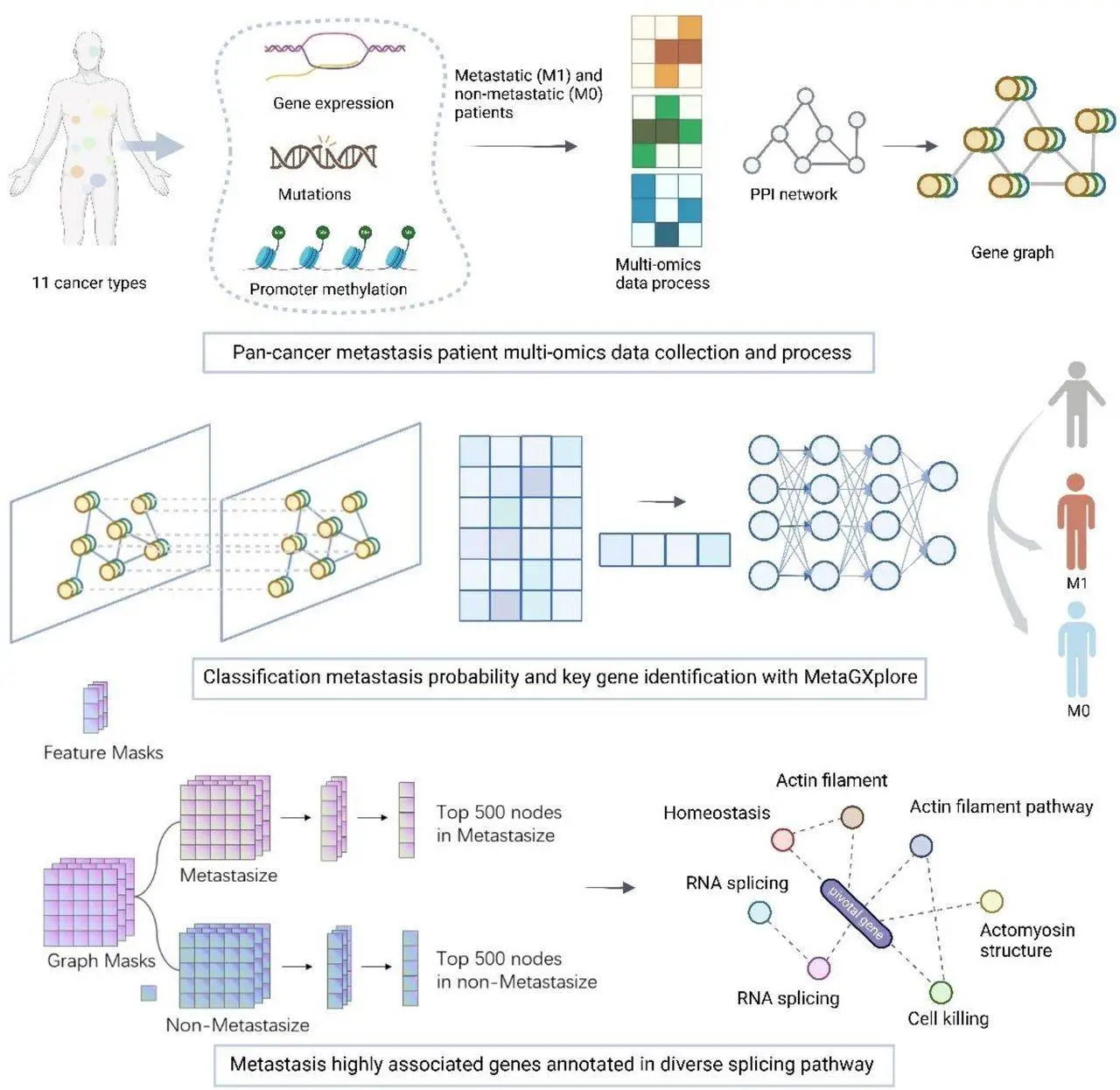
A significant obstacle to the clinical prognosis is tumor metastasis, or the spread of cancer cells from the original tumor to other locations. The current approaches to predicting and detecting tumor metastasis are inefficient and unclear since they rely on laborious tests and subjective clinical judgments. For treatment interventions to be successful and for patients to survive, accurate tumor metastasis likelihood prediction is essential. Finding important genes may yield useful information for locating biomarkers unique to each tumor. Using a K-fold cross-validation process, the researchers trained the classification model, and GNNExplainer was used to interpret the projected outcomes. By assessing the model, graph structure contribution, and multi-omics data, MetaGXplore’s efficacy was proven. Lastly, enrichment analysis of differentially expressed genes and pathways was carried out to investigate the biological roles of significant differential genes and the connections between important pathways.
Introduction
One of the most important indicators of malignancy is cancer metastasis, which is the spread of cancer cells from the main tumor to distant organs. This procedure presents a significant problem for the therapy of cancer since it frequently indicates advanced tumor growth and has a detrimental effect on patient survival. The histological exams, imaging modalities, and clinical findings that are the foundation of current metastasis identification approaches may not be sensitive enough to identify micrometastases or forecast the likelihood of metastatic spread. Innovative instruments are therefore required for the quick and easy evaluation of tumor metastatic risk.
AI in Health
Artificial intelligence (AI) has become a transformative approach in disease research and clinical practice, particularly in disease research. Numerous applications have demonstrated the great potential of AI-based predictive models that use multi-omics data. The number of molecular traits that can be utilized as features and tools to understand pathophysiology has greatly expanded with the development of high-throughput sequencing technology and multi-omics sequencing. Patients’ molecular features vary due to the intrinsic heterogeneity of cancer. Better predictive treatment methods, deeper insights, and better tumor metastasis risk prediction are made possible by integrating and translating these patient genetic traits into patient-specific networks.
The enormous dimensionality of data points from multi-omics datasets that represent unique patient traits presents a problem for current techniques, causing instability in feature selection across datasets. Furthermore, it is crucial to interpret deep learning model predictions in a genetic context, particularly in light of legal obligations such as the General Data Protection Regulation (GDPR) of the European Union. Thus, a key component of improving trust in AI model predictions and comprehending the principles behind tumor metastasis is deep neural network interpretability.
Understanding MetaGXplore
MetaGXplore is a deep-learning artificial intelligence model that predicts metastasis using multi-omics pan-cancer data and Graph Convolutional Networks (GCNs). After training on 754 data spanning 11 cancer types, the model outperformed transformers and Multi-Layer Perceptron (MLP). The Graph Mask and Feature Mask functions of GNNExplainer were utilized to improve interpretability. The underlying workings of the model were clarified, and important genes linked to metastasis were found using this approach. Pathway enrichment analysis made understanding gene relationships and activities within Protein-Protein Interaction (PPI) networks possible. In addition to providing useful gene sets for the following studies on tumor metastasis, this work advances AI and multi-omics data in clinical treatment strategy selection.
MetaGXplore sheds light on metastasis pivotal genes.
GNNExplainer is a prediction framework that ranks genes for metastatic and non-metastatic patient prognostication using graph mask and feature mask approaches. Many biological activities, such as RNA splicing, actin filament organization, anatomical structure homeostasis, cell death, and RNA splicing regulation, are associated with the top 500 genes. These routes frequently cross paths with mechanisms of cancer metastasis, including invasion, migration, and environmental adaptability of cells.
T cell-mediated cytotoxicity, antigen processing, MHC class 1b presentation, and metabolic processes such as ceramide metabolism, sphingomyelin, and sphingoglycolipid metabolism are among the noteworthy pathways that have been highlighted. Significant enrichment in RNA splicing was seen for multiple genes, and somatic mutations in pre-mRNA splicing factors were found to indicate early genetic events and frequently occurring acquired mutations in myelodysplastic syndromes and associated myeloid tumors.
The interpretability study of MetaGXplore showed a noteworthy enrichment in new pathways linked to patients who were not metastatic. These pathways—which support tumor cell self-clearance, maintain cellular structural integrity, and boost anti-tumor immune responses—may obstruct or prevent the spread of metastatic disease. These pathways include macroautophagy, cellular component assembly, intermediate filament cytoskeleton organization, and positive cell-killing regulation. This finding offers a theoretical framework for comprehending the molecular orchestration of metastasis and developing targeted therapeutics for patients who are not metastatic.
Conclusion
Researchers have developed a novel deep-learning model to predict the chance of metastatic development in patients suffering from different types of cancer. Using a framework that is easy to understand, scientists have identified a critical group of genes that play a significant role in prediction and may be connected to the spread of cancer. With a predictive method to assess the metastatic risk for each patient, the results provide healthcare professionals with an invaluable resource. Enriching the knowledge of this complex process and elucidating this gene set also provides insights into the underlying molecular mechanisms controlling metastasis.
Article Source: Reference Paper | Access data and model for this article are listed on GitHub.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.