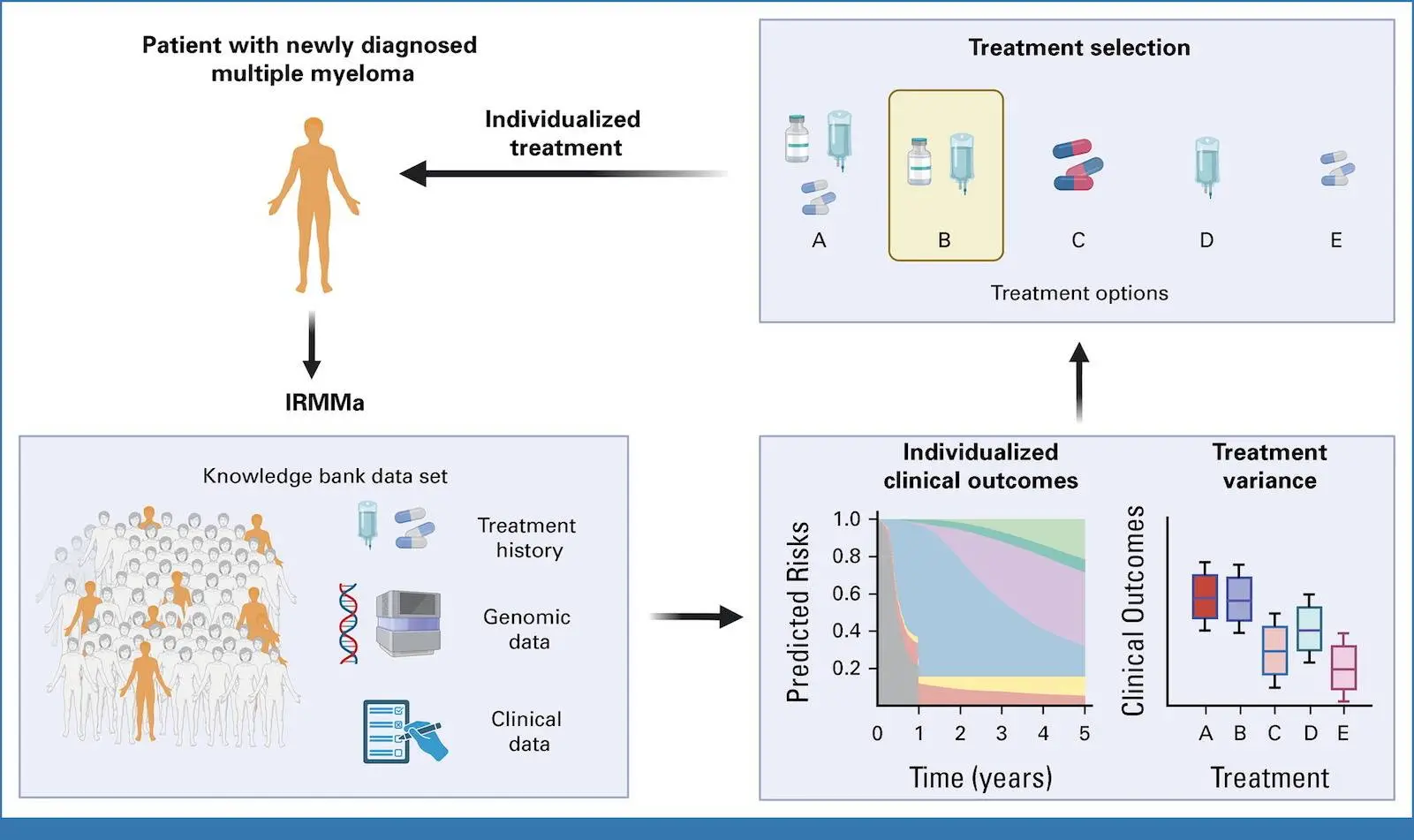
Patients with recently diagnosed non-differentiated multiple myeloma (NDMM) have a wide range of outcomes, with overall survival (OS) spanning from several months to more than ten years. In this work, the researchers gathered a significant training set and a validation set of NDMM patients with clinical, demographic, genomic, and treatment data for the construction of the genomic classification and the prediction model for individualized risk in NDMM (IRMMa). Twelve categories were found using a thorough catalog of genetic factors to predict IRMMa. This model corrected for time-dependent variables such as high-dose melphalan followed by autologous stem-cell transplantation (HDM-ASCT) by integrating clinical, genomic, and therapeutic data. With 256 patients participating in the GMMG-HD6 clinical trial, the model was validated, and it was found that treatment approaches had a significant impact on each patient’s risk in each of the 12 genetic categories.
Introduction
The development of novel treatment drugs has improved the clinical results for newly diagnosed multiple myeloma (NDMM). There is still a lot of variation, though, with some patients getting very little from more advanced treatments and others living disease-free for decades. The goal of suggested changes to the International Staging System (ISS) based on fluorescence in situ hybridization (FISH) is to improve the definition of risk in NDMM. However, considerations including patient-to-patient variability, therapy exclusion, and the widespread disregard for time-dependent and genetic traits that are prognostically significant limit their utility in clinical practice. Although whole-genome, whole-exome, and targeted sequencing studies have found genomic characteristics, FISH and gene expression profiling (GEP) models continue to be the primary methods used in the clinical classification of multiple myeloma (MM). There are difficulties in creating reliable methods for classification and grouping that account for the co-occurrence of several genetic characteristics. Approaches to large-scale data integration may be able to distinguish between different patient groups that benefit from different treatments.
Understanding IRMMa
The goal of the study was to use therapeutic data from a sizable training and validation sample of patients to create a thorough genomic categorization of NDMM. As a result, the first personalized prediction model with mixed clinical and genetic data was developed. After a median follow-up of 43 months, the Eastern Cooperative Oncology Group developed IRMMa to predict individualized risk for OS (death) and EFS (predicted risk of progression). The model outperformed other prognostic models, such as ISS, revised (R)-ISS, and R2-ISS, in terms of accuracy. A number of genomic characteristics, including 1q21 gain/amp, TP53 loss, and 1p deletions, greatly increased model accuracy. The most significant clinical characteristics for model accuracy were age and ISS; the effects of sex, race, Eastern Cooperative Oncology Group, and lactate dehydrogenase (LDH) were less pronounced. One important factor influencing risk was the choice of first-line therapy, indicating that efficient treatments may alter the risk related to genetic and clinical factors.
The researchers were able to incorporate and measure the influence of time-dependent elements like HDM-ASCT and maintenance/continuous treatment because IRMMa was designed as a multistate model. Compared to earlier models, this represents a significant methodologic advancement that makes it possible to assess and make corrections to the clinical impact of these two post-induction treatments. According to the most recent research, phase II EFS was significantly impacted by HDM-ASCT and maintenance/continuous treatment, but OS was less affected. All things considered, these findings show how crucial it is to take therapy and genetic characteristics into account when estimating the OS and EFS of NDMM patients, respectively. For instance, a patient with a low-risk genetic profile might have a brief extension of survival (EFS) if they are not exposed to a treatment that works for their specific illness subtype. However, the effects of different and maybe more successful following therapy may not affect OS for the same patient. On the other hand, a patient with a high-risk genetic profile typically had a short EFS and OS and was resistant to most treatments.
In the GMMG-HD6 experiment, 256 patients underwent testing of the IRMMa model, which demonstrated superior accuracy for EFS and OS in comparison to other models. High agreement between predicted risks and observed results was also demonstrated when the model was employed as a knowledge bank to predict outcomes in the GMMG-HD6 cohort.
IRMMa Advantage over other Prognostic Models
The capacity of NDMM treatment prognostic models, such as R-ISS and R2-ISS, to forecast patient-level individual risk is constrained. However, there are significant benefits over earlier models with IRMMa, a prediction model that combines genetic classification, clinical, demographic, and therapy data. By combining essential elements from deep neural networks and genetic classification, this model, called IRMMa, offers a more precise and individualized method of addressing patient outcomes. The advantages of IRMMa over other prognostic models are:
- When clinical, demographic, and therapeutic factors are taken into account, IRMMa incorporates genetic traits that have been chosen based on their prognostic significance. Expanded genomic characterization in NDMM prognostication is necessary, as evidenced by the significant improvement in IRMMa’s capacity to identify primary refractory and early progressive patients, as well as the boost in accuracy for OS that comes from the inclusion of 20 very relevant genomic markers.
- With the help of IRMMa, the risk of death or advancement for a specific NDMM patient can be estimated, taking into account consolidation and treatment plans. Although it has been demonstrated that HDM-ASCT and maintenance/continuous treatment greatly enhance EFS, their time-dependent character has prevented them from ever being taken into account in the creation of earlier prognostic models (R-ISS and R2-ISS). These characteristics were made possible by the IRMMa multistate design, which increased the overall accuracy of EFS. Moreover, a crucial tool for choosing the best course of action and preventing overtreatment in situations where it offers little to no benefit is the capacity to record each patient’s unique treatment variance. Relevantly, IRMMa may be useful in determining which patients benefit from HDM-ASCT and which do not. A number of phase III randomized trials have investigated the potential benefits of HDM-ASCT as a consolidation approach following IMIDs and/or PIs. HDM-ASCT has generally been linked to an advantage in PFS but not in OS, according to the majority of studies. These findings bring up crucial clinical issues regarding the best ways to counsel NDMM patients, especially when new, potent immunotherapies become available in the future.
- Age, therapy, and ISS are required features of the treatment model IRMMa. Even with a lower percentage of concordance, IRMMa can produce estimates in the absence of genomic data; it can outperform R2-ISS, R-ISS, and ISS, offering a chance to enhance predictions in the absence of complete genomics.
Limitations of IRMMa
- IRMMa was constructed using genomic data from a single bone marrow site and does not take into account the potential impact of genomic drivers at distinct anatomic sites (i.e., spatial heterogeneity).
- The sample size utilized in the training set was less than the one used to generate R-ISS and R2-ISS. Future integration of liquid biopsy and bone marrow techniques may enhance the resolution and performance of IRMMa even more.
- Lastly, because these data are not currently available for sufficiently large cohorts, the present IRMMa model is unable to provide estimates for novel drugs (such as antiCD38 antibodies) and unique time-dependent properties (such as minimal residual disease).
Conclusion
In an effort to increase therapy sensitivity, the study focuses on NDMM patients and genomic causes. The work finds important genetic drivers and suggests an extensive genomic classification system by analyzing a wide range of data sets. By capturing the variety among molecular subgroups, this method improves our comprehension of treatment sensitivity and clinical heterogeneity. Compared to prior efforts, the study offers advantages like a larger sample size, a more thorough analytical approach, and an emphasis on genetic patterns of co-occurrence. With NDMM patients’ heterogeneity currently oversimplified, IRMMa offers a novel chance to better examine it and enhance our comprehension of the results of both past and future therapeutic studies.
Article source: Reference Paper | Reference Article
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.