Researchers from Sorbonne University and Dauphine University, France, introduced MetagenBERT, a Transformer-based framework for disease prediction directly from raw metagenomic DNA without relying on taxonomic annotations. By achieving strong performance across multiple gut microbiome datasets using fewer reads, the study highlights a scalable, annotation-free approach with potential for robust, cross-cohort metagenomic disease prediction.
Why Current Metagenomic Methods Are Limited
Metagenomic analysis commonly relies on abundance-based representations, but this approach has a problem: it depends on how detailed our taxonomic knowledge is. It also relies on databases that are incomplete, and it simplifies complex sequences down to just counts of species, which means we can miss out on important functional information available.
Although these depictions have been employed in deep learning and machine learning models for disease categorization, their scalability is constrained by evolving reference databases. Though genomic large language models allow transfer learning for a range of activities and learn sequence patterns straight from DNA, their performance is erratic, and long-range dependencies prove difficult. Though they are still task-specific and lack a general, independent reference foundation for accurate metagenomic representation, recent metagenomic-specific large language models (LLMs) improve tasks like pathogen detection and annotation.
End-to-End Metagenome Representation Using MetagenBERT
MetagenBERT is a deep learning framework that accepts raw Sequencing rates to generate metagenome embeddings. It employs genomic language models to extract biologically meaningful patterns from DNA sequences and enables metagenomic representation and classification without dependency on annotations or reference databases.
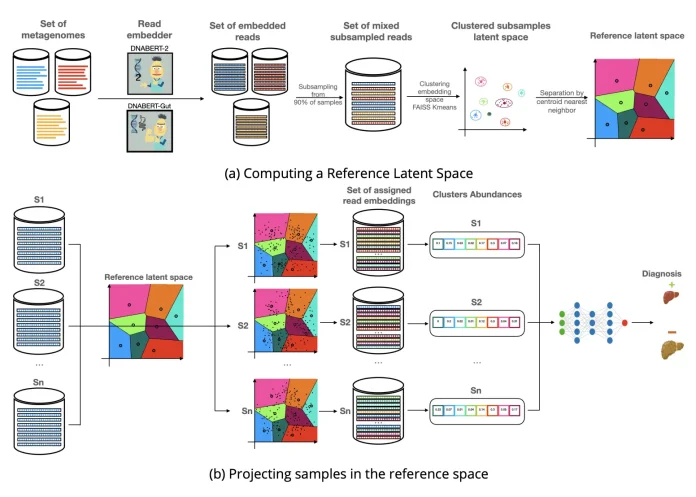
A metagenomic sample has millions of short DNA reads stored in FASTA/FASTAQ files. MetagenBERT uses two main stages to produce end-to-end embedding, read embedding, and metagenome embedding. In the first stage, a genomic large language model is used to convert sequencing reads into numerical vectors. In the second stage, these read-level embeddings are aggregated, and a fixed-length vector is generated representing the entire metagenome that can be used for downstream disease classification.
Further, researchers analyzed 5 commonly used shotgun metagenomic gut microbiome data sets containing case-control samples, which include liver cirrhosis, colorectal cancer (CRC), inflammatory bowel disease (IBD), obesity, and type 2 diabetes (T2D). These data sets have complex, feature-rich data and limited sample size characteristics of metagenomic studies. Also, the metacaCardis dataset, which focuses on cardiometabolic disorders in European populations, was used to train the model quality control was performed by using fastp for benchmark datasets.
DNABERT-2, a general-purpose genomic language model trained on DNA sequences, was used to read embeddings, and DNABERT-MS was trained on simulated Metagenomic reads using masked language modeling to better capture gut-specific patterns. Further, both models were evaluated using taxonomic classification and reconstruction loss, demonstrating that DNABERT-MS is slightly better than DNABERT-2 in capturing gut microbiome-specific features.
Mean pooling was applied to produce one embedding per read, and only 10% of reads per sample were used to reduce computational cost while maintaining the performance; hence, each metagenome was represented by millions of 768-dimensional read embeddings. Further, to summarize all of this, large data clustering was performed using a global K-means algorithm implemented with FAISS for efficient GPU computation, so each metagenome was then represented as a cluster abundance vector by assigning reads to the nearest clusters. A LASSO classifier was used for disease classification based on species abundance cluster-based embedding and their combinations.
MetagenBERT gave strong disease prediction performance especially for T2D, CRC and IBD wear cluster based embeddings out performed the traditional species-abundance approach. Combining cluster and species information drastically improved results, indicating that they capture complementary biological signals. MetagenBERT outperformed most models for IBD, and type 2 diabetes also performs slightly worse than EnsDeepDP for obesity and colorectal cancer. Lastly, sub-sampling experiments showed that using only 10% of reads produces results similar to using 100% of reads. Cluster analysis indicated that case and control samples shared a common overall structure with disease-specific differences in certain regions.
Conclusion
MetagenBERT-Glob overcomes significant metagenomics problems without relying on species annotations or inadequate reference databases by creating sample-level embeddings straight from raw DNA readings. By embedding and clustering millions of reads, it finds microbiome patterns that improve conventional species-based techniques as well as the prediction of disease.
Performing well over multiple datasets shows the approach’s promise as a universal tool. Still, it has downsides, including the necessity of a lot of processing power, the challenge of deciphering clusters, and the chance of unequal results with tiny or biased datasets. Larger, more diverse datasets and better models help to reduce these constraints, hence extending MetagenBERT-Glob’s use.
Article Source: Reference Paper | Code Availability: GitHub.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Important Note: bioRxiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Jainab Shaikh is a postgraduate in Biotechnology with a strong interest in understanding how research translates into real-world innovation. Her areas of focus include biosensors, bioinformatics, and sustainable biotechnological applications. She is passionate about exploring recent scientific advancements and communicating them through clear, engaging, and accessible content. Her work particularly emphasizes research-driven narratives in healthcare, biotechnology, skincare science, and emerging life science innovations.