Researchers from École Polytechnique Fédérale de Lausanne (EPFL) and the National Centre of Competence in Research (NCCR) Catalysis, Switzerland, introduced DynaMate, an autonomous AI system that can fully set up, run, and analyze molecular dynamics simulations for proteins and protein-ligand complexes. DynaMate employs a multi-agent large language model architecture to plan simulation workflows, prepare molecular systems, and interpret results, while also correcting errors that typically require expert human intervention. DynaMate lowers technical barriers and enables more scalable, efficient molecular modeling for drug discovery and protein engineering.
Overview
Molecular Dynamics Simulations rely on mathematical models that describe how atoms and molecules interact to predict how candidate molecules bind to proteins over time based on classical physics laws. Despite their utility, MD simulations are hard to set up, especially as a beginner, due to their error-prone and time-consuming nature.
This problem can be solved by automating the entire MD simulation such that it does not require any human intervention. Researchers have developed one such modular multi-agent AI framework that automatically plans, executes, troubleshoots, and analyzes results on its own once the goal is given.
Why Molecular Dynamics Remains Technically Complex
In molecular dynamics, as the name suggests, each atom is treated as a particle that is allowed to interact with proteins, and the forces between atoms are calculated using a force field over a tiny time period.
A force field is a mathematical model that describes how atoms interact, considering bond stretching, angle bending, torsion, non-bonding interactions like electrostatic and van der Waals, and many more constraints. This helps researchers understand protein structure and flexibility, protein ligand binding, and predict this bond’s stability.
The conformations generated by MD are used to calculate the binding energy, which in turn judges how strongly a ligand binds to a protein.
Traditionally, setting up MD simulations required expert knowledge, as it’s a multistep process.
It involves retrieval and cleaning the structures, assigning protonation states, solvating the system, choosing a force field, and, most tediously of all, parameterizing the ligands.
Each step requires domain knowledge and is dependent on the previous one; hence, the process is prone to errors, especially when it comes to ligand parameterization. Small mistakes can cause simulation crashes and invalid results.
To make MD simulation easier to operate, earlier automation tools like CHAPERONg and PyAutoFEP were aimed at reducing manual intervention. However, they were rigid, meaning that any new molecule type, force field, or simulation engine required manual re-engineering by an expert.
Automation in Molecular Dynamics Simulations
A large language model normally just answers questions or generates texts. An agentic LLM goes further; it can act like an agent, meaning it doesn’t just respond but can also plan, decide, and execute multistep tasks on its own.
Think of it as an AI agent that can:
- Understand your request
- Break it into steps
- Use external tools/software to carry out those steps
- Adapt if something goes wrong
When it comes to MD simulations, agentic LLMs can automate the error-prone steps explained earlier in the blog, reducing the manual effort. They can plan workflows, execute parameterization, and recover from errors dynamically. Hence, they can transform MD from a manual, expert-heavy process to a flexible, adaptive pipeline useful in drug discovery.
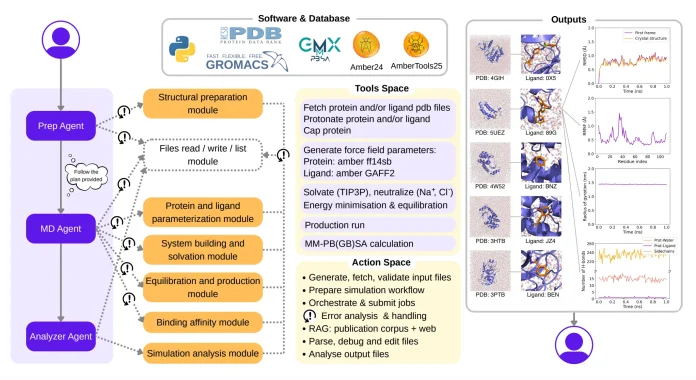
DynaMate: An Overview of Its Design and Workflow
DynaMate is an agentic LLM framework built specifically for molecular dynamics simulations. It uses a multi-agent design. Which means instead of one big agent, DynaMate uses multiple specialized agents that coordinate together. Such as one agent handles high-level reasoning (planning, parameter selection, etc.), another handles low-level execution (simulation setup, file handling, and running MD engines). This makes workflows more reliable and adaptable.
DynaMate can also choose the right external tools (eg, GROMACS, OpenMM) depending on the task, instead of being locked to one engine. It can pull information from external sources like web searches or existing literature to make smarter decisions about parameters, force fields, or protonation states.
What makes DynaMate hold up to its name is that if a simulation crashes or produces unstable results, DynaMate can diagnose the issue and adjust settings automatically, rather than requiring human intervention. The authors tested Dynamate on several protein and protein-ligand systems and found that it could successfully complete full simulations, and when used to calculate binding free energies, its results showed values similar to those of experimental data.
Conclusion
Setting up and running MD simulations is technically complex, particularly when it comes to small ligand molecules. Agentic LLMs can autonomously plan and execute complete MD frameworks, overcoming complexities and demonstrating their potential to scale molecular modeling for future biomolecular and drug design applications.
Article Source: Reference Paper | Code availability: GitHub
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Saniya is a graduating Chemistry student at Amity University Mumbai with a strong interest in computational chemistry, cheminformatics, and AI/ML applications in healthcare. She aspires to pursue a career as a researcher, computational chemist, or AI/ML engineer. Through her writing, she aims to make complex scientific concepts accessible to a broad audience and support informed decision-making in healthcare.















