Even though SMILES and molecular graphs have been used in earlier research, drug reactions in biological systems, such as genes and proteins, are frequently disregarded in the de novo production of hit-like compounds. In this study, scientists from Nagoya University and Kyushu Institute of Technology, Japan, provide Gx2Mol, a deep generative model that creates molecular structures with desired behaviors for any target protein by using gene expression profiles. To discover the latent feature distribution of the gene expression profiles, the approach uses a variational autoencoder as a feature extractor. In order to generate syntactically valid SMILES strings that meet the feature criteria of the gene expression profile that the feature extractor has extracted, a long short-term memory is then used as the chemical generator. The suggested Gx2Mol model can generate novel compounds with possible bioactivities and drug-like qualities, as shown by experimental findings and case studies.
Introduction
Exploring compounds that have therapeutic benefits, like anticancer medications, is a risky and expensive process. Drug development takes more than 12 years and costs more than $1.3 billion, and the failure rate is still high despite rigorous premarket testing. The process of looking for possible medication candidate molecules that have the ability to interact with therapeutic target proteins is known as hit identification. For hit identification, high-throughput screening of extensive chemical compound libraries using biological assays is frequently carried out; nevertheless, this is costly.
Computational techniques like virtual screening and de novo molecule generation provide an alternative to hit identification. Virtual screening uses docking simulation or ligand-based similarity search to find hit-like molecules from chemical databases at a low cost. The goal of de novo molecular production is to create novel compounds that resemble existing ligands or have specific chemical characteristics. Generative models for de novo molecule production have been developed using deep learning and artificial intelligence.
Understanding Gx2Mol
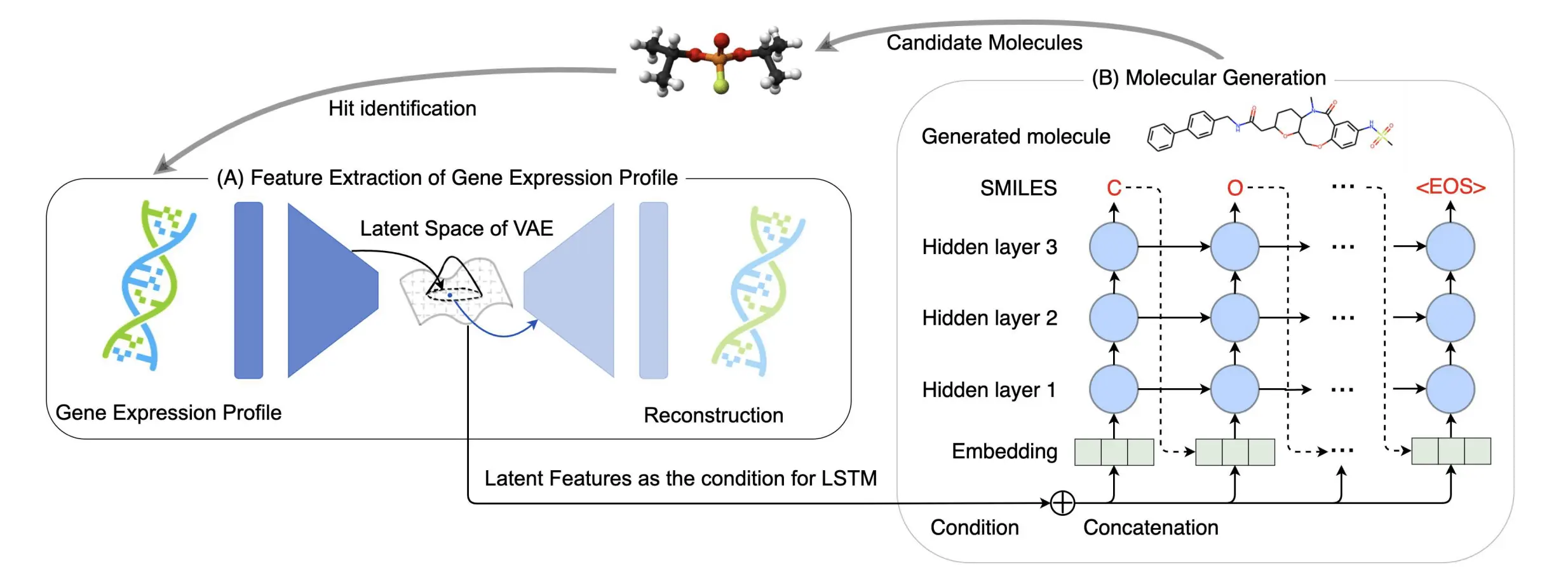
The research introduces Gx2Mol, a deep generative model that analyzes omics data and creates novel drug structures using a VAE model as a feature extractor. In order to create synthetically valid SMILES strings that meet the feature criteria of the gene expression profile that the VAE-based feature extractor has extracted, the LSTM model is employed as a chemical generator. Gene expression profile features are used as training conditions for the LSTM model, which directs the model to produce compounds linked to the target gene expression profile. Since the suggested approach produces new compounds with possible bioactivities and drug-like qualities that can be used for additional structure optimization, the main contributions are a fresh idea, a succinct model, and higher performance.
The architecture of the Gx2Mol model
There are two primary parts to the suggested Gx2Mol: the generator, also known as the conditional LSTM model, is used to create hit-like molecules based on the features of the gene expression profiles that have been retrieved using the feature extractor, also known as the VAE model.
To produce hit-like compounds from gene expression profiles—which frequently struggle with noise and redundant information—VAE models offer a calculated answer. By capturing the underlying structure of high-dimensional data and obtaining a lower-dimensional representation, generative models, or VAEs, can comprehend its intricacies. Gene expression profile noise and redundancy problems are successfully navigated and mitigated by this approach. High-dimensional gene expression profiles can be made less dimensional using VAEs, which develop a meaningful and compressed representation in the latent space while capturing key characteristics. Through the modeling of the data’s underlying distribution, noise removal, and the capture of inherent patterns, they also aid in noise reduction. It is possible to think of the latent space that VAEs learn as a collection of features that have been retrieved and represent crucial details in gene expression profiles.
Limitations and Future Work
There is one major restriction to this study, i.e., there may be a limit on the variety of molecules produced when utilizing LSTMs as generators because they are commonly used in auto-regressive generation activities, in which the token at the subsequent time step is generated based on the token at the current time step.
In subsequent studies, researchers want to investigate methods for improving the variety of molecule creation in the suggested Gx2Mol model. Moreover, the Gx2Mol model is intended to be integrated into useful AI systems to help scientists create a variety of drug-candidate hit-like molecules that are suited for different illnesses. The advantages of the Gx2Mol model should be capitalized upon by this integration, which should also help develop drug discovery procedures.
Conclusion
The Gx2Mol model is a deep learning method that uses gene expression profiles to produce possible chemical structures of molecules that resemble hits. Following feature extraction using a VAE during the training phase, syntactically valid SMILES strings are produced. Hit-like molecules are generated by the VAE encoder, which is the only feature extractor in the generation phase. In particular, the approach has been useful in producing hit-like compounds from gene expression profiles in therapeutic treatments for Alzheimer’s disease, stress dermatitis, and stomach cancer.
Article Source: Reference Paper | Source code is available freely on GitHub.
Disclaimer:
The research discussed in this article was conducted and published by the authors of the referenced paper. CBIRT has no involvement in the research itself. This article is intended solely to raise awareness about recent developments and does not claim authorship or endorsement of the research.
Important Note: arXiv releases preprints that have not yet undergone peer review. As a result, it is important to note that these papers should not be considered conclusive evidence, nor should they be used to direct clinical practice or influence health-related behavior. It is also important to understand that the information presented in these papers is not yet considered established or confirmed.
Follow Us!
Learn More:
Deotima is a consulting scientific content writing intern at CBIRT. Currently she's pursuing Master's in Bioinformatics at Maulana Abul Kalam Azad University of Technology. As an emerging scientific writer, she is eager to apply her expertise in making intricate scientific concepts comprehensible to individuals from diverse backgrounds. Deotima harbors a particular passion for Structural Bioinformatics and Molecular Dynamics.