Understanding complex cellular behaviors quantitatively is essential for gaining in-depth insights into cell biology. Tracking the movement and interactions (movies) of cells over time gives a lot of insights into cellular mechanisms, but analyzing this data presents significant challenges. Traditional methods often involve cumbersome software and require considerable computational resources, limiting the ability of researchers to derive meaningful conclusions efficiently. In response to these challenges, the development of CellTracksColab by researchers from the Instituto Gulbenkian de Ciência, Portugal, the Åbo Akademi University, Turku, Finland, the University of Turku, the Université Paris Cité, and the University College London, marks a significant advancement, offering a streamlined and accessible platform that enables the compilation, analysis, and exploration of cell tracking data with unprecedented ease and flexibility.
Why do we need CellTracksColab?
Cell tracking data is very important for many biological studies, including cancer research, immunology, and developmental biology. As imaging technologies have advanced, the volume of data generated in these studies has increased exponentially. However, the tools available for analyzing this data have not kept pace, often requiring specialized knowledge and infrastructure that are not universally available. This disconnect between data generation and analysis capabilities has created a bottleneck in research, limiting the speed and scope of scientific discoveries.
Enter CellTracksColab: with data gathering, standardization, and learned methods of analysis all in one convenient place- with a friendly portal- the platform of CellTracksColab allows the researcher to concentrate on the scientific problems at stake rather than the methodical problems of data management! Why is it so great? The design of the platform goes to where the cell biology community sees the need to provide both powerful tools and accessibility. As a bonus, it can be used with Jupyter Notebooks, which can easily be used by many researchers and programmers.
Key Features of CellTracksColab
- Seamless Integration with Jupyter Notebooks
Not only did the CellTracksColab, one of the key features, integrate the Jupyter notebooks, but the latter is also a crucial tool amongst the scientific community for the analysis and visualization of data. Jupyter Notebooks, being an excellent interactive computing environment, provide a space to run code, house data, and document work- all within one common place- whose necessity for undertaking science cannot be overemphasized. CellTracksColab applies this environment, making it available within a notebook for the user to compile and analyze within one workflow. This also serves reproducibility, which is one of the backbones of scientific research. Through Jupyter notebooks, a researcher can document the entire process of analysis to make them easily reproducible by others. It proves very relevant in cell biology because, with it, the experimental conditions in use and the steps taken in data processing can have significant effects on the outcomes of a research study.
- Accessibility and Collaborating with Jupyter Notebooks in the Cloud
In addition to the local implementation, CellTracksColab is deployed on cloud-based platforms, including Google Colab. This increases access to the platform for researchers who do not have good access to facilities with high-performance computational resources. The possibility of running complex analyses over cloud infrastructure starts to democratize advanced data processing tools and will be a significant opportunity to involve more people in leading-edge research in cell biology.
Second, collaboration is made possible by cloud-based execution, which allows many users to execute on the same dataset concurrently. For this reason, it is easier to share findings and conduct projects with several other people. This comes in handy in larger interdisciplinary teams where each member might contribute a particular analytical part. CellTracksColab has thus allowed a harmonized, generous, and productive research environment on a common platform.
- Comprehensive Analytical Tools
Central to CellTracksColab, one will find an array of analysis tools to make sure data on cell tracking conveys insight. These are not data processing tools; the user will find included high-level analytics in order to measure the behavior of cells in all sorts of different ways. For example, cell speed, cell displacement, and cell trajectory taken over time will present a very detailed view of how cells are moving and interacting across an experiment. These key metrics are very important when trying to understand processes like cell migration, division, and differentiation that are so common in a host of biological research topics. In addition, CellTracksColab enables the quantification of certain cellular events- such as mitosis or apoptosis- via the identification of specific events in time. This reduces any possible human mistakes and guarantees that no time lags are instigated in the process of the screening course. Due to the fast change of needs and priorities in the scientific research community, the new analytical tools and algorithms must always be incorporated in a flexible way. Extensibility is also intimately associated with keeping CellTracksColab current with respect to research on cell motility tracking.
- Customizable and Informative Visualizations
Visualization is one of the most important parts of data analysis, and it is particularly important in this field where things are very visually oriented, such as in cell biology.
CellTracksColab supports multiple areas of visualization to supplement the needs of the varied researchers associated with it, such as 2D and 3D plots, heat maps, and maps of trajectories, which can all be customized for specific aspects of the data. In addition to being informative, these visualizations could explain complex results to an extended audience very effectively, making them understand even if they are not experts in data interpretation.
- Open Source and community-driven
As such, an open-source platform, CellTracksColab, steamrolls those efforts of transparency and collaborative platforms in the scientific community. Making the platform freely available encourages the contribution of researchers to its continuous development. Thereby using community-driven approaches, it is assured that the tool is responsive to the needs of the users with added constant features and improvements.
Furthermore, the collaborative and open-source nature of CellTracksColab entails added educational value to a student or early-career researcher. Already, with this infrastructure, students can interact with the cutting-edge tools of analysis of cell tracking; this way, the system can train the next generation of cell biologists and prepare this ever-growing community for the new challenges in modern research.
Results
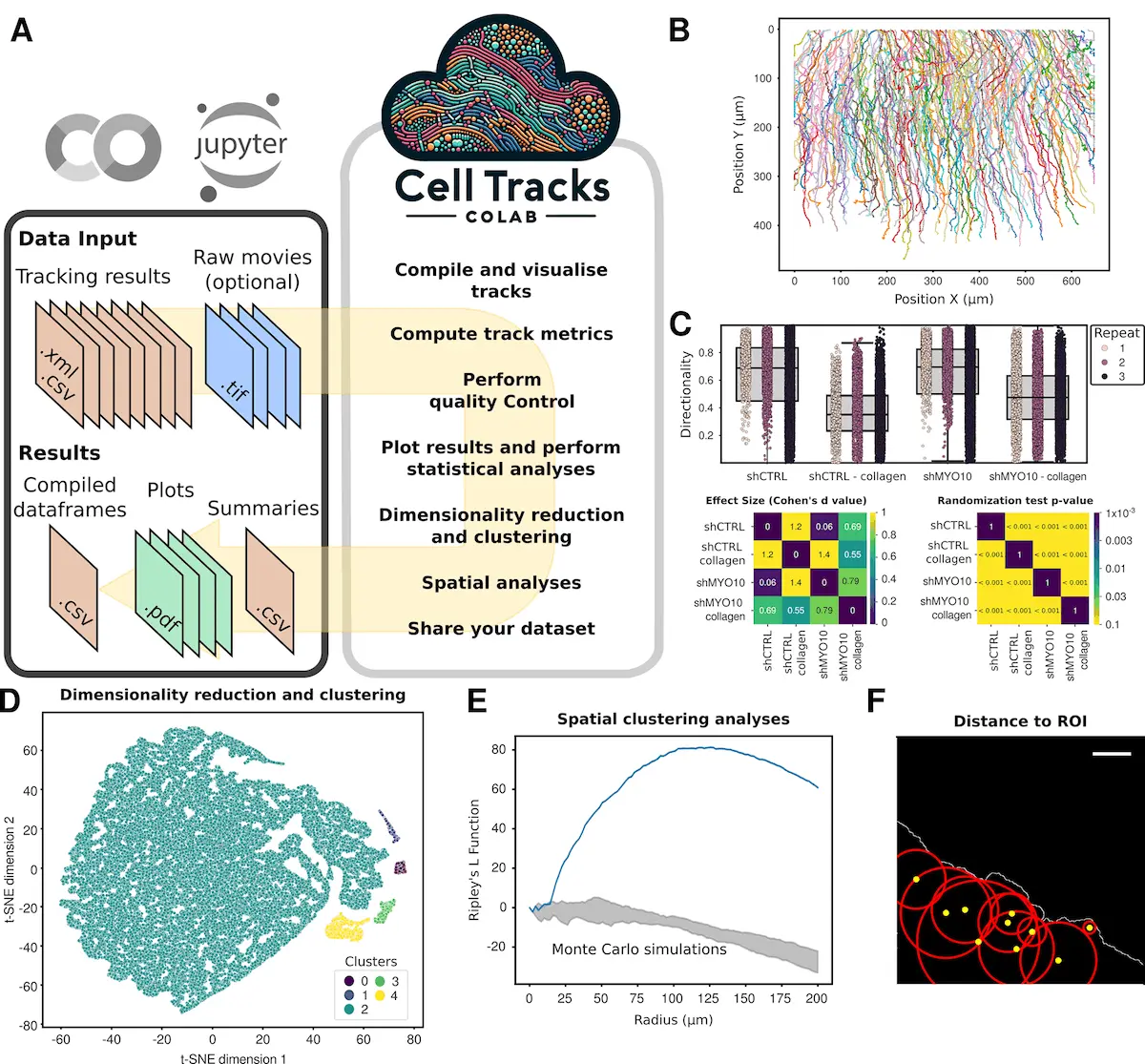
When these datasets of T cells migrating on VCAM (Vascular Cell Adhesion Molecule 1) or ICAM (Intercellular Adhesion Molecule 1) were reanalyzed using CellTracksColab, we found that T cells on ICAM migrate faster and more directionally.
UMAP (a visualization technique called Uniform Manifold Approximation and Projection, which gives a dot plot “reduced” from each other according to certain logical features) in combination with clustering shows evident distribution patterns and some behaviors resembling VCAM on ICAM. The data obtained using this technology showed that MYO10-silenced cells had a slower yet more directed movement under the collagen gel in a dataset involving cancer cells. In another study, it was shown that MYO10WT-associated filopodia were stable and moved slower than those associated with the MYO10 variants in U20S cells.
Conclusion
CellTracksColab is such a large step toward the analysis of cell data on the track, combining the strength of Python, Jupyter Notebooks, and the facility of cloud computing. In conjunction with a range of tools for seamless analysis and an intuitive interface, this definitely places CellTracksColab among the necessary platforms for all researchers at work in cell biology. By virtue of its ability to facilitate and improve the analysis process, there is a chance for CellTracksColab to enormously contribute to bringing new revelations closer to the researchers and to understand the “movie” concerning the dynamics of the cell.
Article Source: Reference Paper | CellTracksColab is available to the broader scientific community on GitHub.
Follow Us!
Learn More:
Neermita Bhattacharya is a consulting Scientific Content Writing Intern at CBIRT. She is pursuing B.Tech in computer science from IIT Jodhpur. She has a niche interest in the amalgamation of biological concepts and computer science and wishes to pursue higher studies in related fields. She has quite a bunch of hobbies- swimming, dancing ballet, playing the violin, guitar, ukulele, singing, drawing and painting, reading novels, playing indie videogames and writing short stories. She is excited to delve deeper into the fields of bioinformatics, genetics and computational biology and possibly help the world through research!